Mean ![]() and Standard Deviation
and Standard Deviation ![]() are computed from a given biopsy
of the input image. The parameters a and b are assigned by the user.
This function will find all objects satisfying the threshold criteria shown below:
are computed from a given biopsy
of the input image. The parameters a and b are assigned by the user.
This function will find all objects satisfying the threshold criteria shown below:
- 1.
- The object must contain all voxels which satisfy the following relationship.
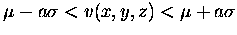
- 2.
- All within the object must be connected to above selected voxels and
should satisfy the following relationship.
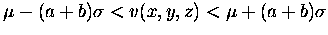 .
.
Note that ![]() and
and ![]() for this function.
for this function.
An 8-bit grayscale volume.
A single biopsy image (obtained using Interseg).
cue_filename = foobar
a_value = 1.0
b_value = 1.0
- 1.
- This function is comparable in run time to the 2-D conn-comp and 3-D conn-comp operations.
- 2.
- A variation of Hysteresis_Threshold function.